Enhancer RNA
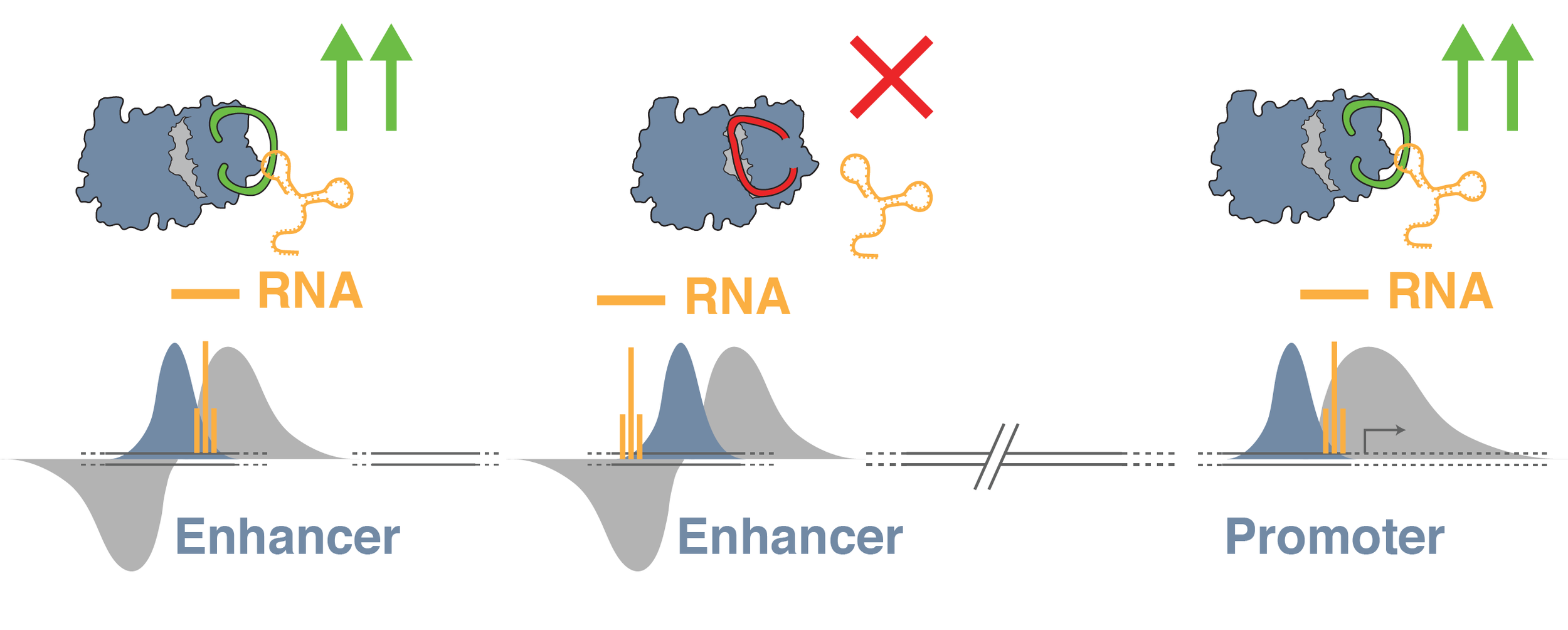
A key candidate for developing diverse enhancer-activity profiles are non-protein coding RNAs, called enhancer RNAs (eRNAs), that are transcribed from active enhancers. Our work has shown how eRNAs can interact with epigenetic-enzymes to stimulate enhancer activity by changing epigenetic modifications on chromatin. As enhancers differ in sequence, this suggests that eRNAs are critical drivers of enhancer-specific activity profiles. Moreover, disease-related mutations at enhancers could fundamentally alter interactions between eRNAs and enzymes to disrupt enhancer activity.
How do eRNAs affect enhancer activity?
As the sequence and structure of eRNAs differs between enhancers, they are prime candidates to drive alternative profiles of enhancer activity to control gene expression.
eRNAs and epigenetic enzymes
Our work demonstrated for the first time that eRNAs could interact with and modulate the catalytic activity of the epigenetic enzyme CBP. eRNAs are transcribed bi-directionally (in opposite directions, sense and antisense from each strand of DNA) from open chromatin at enhancers. Using cross-linking approaches (PAR-CLIP) together with next-generation sequencing, we mapped the binding of eRNAs to CBP across the genome. Our work showed that although binding was independent of eRNA sequence, there was strong enrichment for binding of eRNAs to CBP because CBP is bound to chromatin at enhancers across the genome. This we termed 'locus-specificity' and in fundamentally important for the ability of CBP to bind to eRNAs genome wide, wherever CBP is bound to an enhancer.
CBP binds to eRNAs at enhancers.
Left) UCSC genome browser view of CBP-bound eRNAs. Top; GROseq signal showing bi-directional eRNA transcription; Middle; CBP-bound RNAs from PAR-CLIP data; Bottom; Chromatin-bound CBP from ChIP-seq data.
Right) CBP bound RNAs are enriched at transcribed enhancers: Top; Enrichment for CBP-bound RNAs (PAR-CLIP signal) at sites of CBP chromatin-binding (10th decile = most enriched for CBP); Bottom; Enrichment for CBP-bound RNAs (PAR-CLIP signal) at sites of nascent RNA transcription (GROseq signal, 10th decile = most enriched for nascent RNA).